|
Getting your Trinity Audio player ready...
|
First off, why nanopublications?
Nanopublications complement human-created narratives of scientific knowledge with elementary, machine-actionable, simple and straightforward scientific statements that prompt sharing, finding, accessibility, citability and interoperability.
By making it easier to trace individual findings back to their origin and/or follow-up updates, nanopublications also help to better understand the provenance of scientific data.
With the nanopublication format and workflow, authors make sure that key scientific statements – the ones underpinning their research work – are efficiently communicated in both human-readable and machine-actionable manner in line with FAIR principles. Thus, their contributions to science are better prepared for a reality driven by AI technology.
The machine-actionability of nanopublications is a standard due to each assertion comprising a subject, an object and a predicate (type of relation between the subject and the object), complemented by provenance, authorship and publication information. A unique feature here is that each of the elements is linked to an online resource, such as a controlled vocabulary, ontology or standards.
Now, what’s new?
As a result of the partnership between high-tech startup Knowledge Pixels and open-access scholarly publisher and technology provider Pensoft, authors in Biodiversity Data Journal (BDJ) can make use of three types of nanopublications:
- Nanopublications associated with a manuscript submitted to BDJ. This workflow lets authors add a Nanopublications section within their manuscript while preparing their submission in the ARPHA Writing Tool (AWT). Basically, authors ‘highlight’ and ‘export’ key points from their papers as nanopublications to further ensure the FAIRness of the most important findings from their publications.
- Standalone nanopublication related to any scientific publication, regardless of its author or source. This can be done via the Nanopublications page accessible from the BDJ website. The main advantage of standalone nanopublication is that straightforward scientific statements become available and FAIR early on, and remain ready to be added to a future scholarly paper.
- Nanopublications as annotations to existing scientific publications. This feature is available from several journals published on the ARPHA Platform, including BDJ. By attaching an annotation to the entire paper (via the Nanopublication tab) or a text selection (by first adding an inline comment, then exporting it as a nanopublication), a reader can evaluate and record an opinion about any article using a simple template based on the Citation Typing Ontology (CiTO).
Nanopublications for biodiversity data?
At Biodiversity Data Journal (BDJ), authors can now incorporate nanopublications within their manuscripts to future-proof the most important assertions on biological taxa and organisms or statements about associations of taxa or organisms and their environments.
On top of being shared and archived by means of a traditional research publication in an open-access peer-reviewed journal, scientific statements using the nanopublication format will also remain ‘at the fingertips’ of automated tools that may be the next to come looking for this information, while mining the Web.
Using the nanopublication workflows and templates available at BDJ, biodiversity researchers can share assertions, such as:
- Wolves (Canis lupus) occur in forest habitats;
- A grass snake (Natrix natrix) was observed to eat a tree frog (Hyla arborea);
- Ursus meles is a synonym of Meles meles;
- The nucleotide sequence GU682758 can be used to identify the species Araneus diadematus.
So far, the available biodiversity nanopublication templates cover a range of associations, including those between taxa and individual organisms, as well as between those and their environments and nucleotide sequences.

As a result, those easy-to-digest ‘pixels of knowledge’ can capture and disseminate information about single observations, as well as higher taxonomic ranks.
The novel domain-specific publication format was launched as part of the collaboration between Knowledge Pixels – an innovative startup tech company aiming to revolutionise scientific publishing and knowledge sharing and the open-access scholarly publisher Pensoft.
… so, what exactly is a nanopublication?
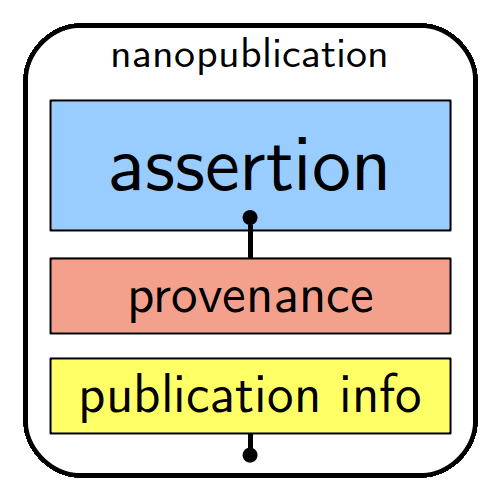
General structure of a nanopublication:
“the smallest unit of publishable information”,
as explained on nanopub.net.
Basically, a nanopublication – unlike a research article – is a tiny snippet of a precise and structured scientific finding (e.g. medication X treats disease Y), which exists as a reusable and cite-able pieces of a growing knowledge graph stored on a decentralised server network in a format that it is readable for humans, but also “understandable” and actionable for computers and their algorithms.
These semantic statements expressed in community-agreed terms, openly available through links to controlled vocabularies, ontologies and standards, are not only freely accessible to everyone in both human-readable and machine-actionable formats, but also easy-to-digest for computer algorithms and AI-powered assistants.
In short, nanopublications allow us to browse and aggregate such findings as part of a complex scientific knowledge graph. Therefore, nanopublications bring us one step closer to the next revolution in scientific publishing, which started with the emergence and increasing adoption of knowledge graphs.
“As pioneers in the semantic open access scientific publishing field for over a decade now, we at Pensoft are deeply engaged with making research work actually available at anyone’s fingertips. What once started as breaking down paywalls to research articles and adding the right hyperlinks in the right places, is time to be built upon,”
said Prof. Lyubomir Penev, founder and CEO at Pensoft, which had published the very first semantically enhanced research article in the biodiversity domain back in 2010 in the ZooKeys journal.
Why are nanopublications necessary?
By letting computer algorithms access published research findings in a structured format, nanopublications allow for the knowledge snippets that they are intended to communicate to be fully understandable and actionable. With nanopublications, each of those fragments of scientific information is interconnected and traceable back to its author(s) and scientific evidence.
By building on shared knowledge representation models, these data become Interoperable (as in the I in FAIR), so that they can be delivered to the right user, at the right time, in the right place , ready to be reused (as per the R in FAIR) in new contexts.
Another issue nanopublications are designed to address is research scrutiny. Today, scientific publications are produced at an unprecedented rate that is unlikely to cease in the years to come, as scholarship embraces the dissemination of early research outputs, including preprints, accepted manuscripts and non-conventional papers.
A network of interlinked nanopublications could also provide a valuable forum for scientists to test, compare, complement and build on each other’s results and approaches to a common scientific problem, while retaining the record of their cooperation each step along the way.
***
We encourage you to try the nanopublications workflow yourself when submitting your next biodiversity paper to Biodiversity Data Journal.
Community feedback on this pilot project and suggestions for additional biodiversity-related nanopublication templates are very welcome!
This Nanopublications for biodiversity workflow was created with a partial support of the European Union’s Horizon 2020 BiCIKL project under grant agreement No 101007492 and in collaboration with Knowledge Pixels AG.The tool uses data and API services of ChecklistBank, Catalogue of Life, GBIF, GenBank/ENA, BOLD, Darwin Core, Environmental Ontology (ENVO), Relation Ontology (RO), NOMEN, ZooBank, Index Fungorum, MycoBank, IPNI, TreatmentBank, and other resources.
***
On the journal website: https://bdj.pensoft.net/, you can find more about the unique features and workflows provided by the Biodiversity Data Journal (BDJ), including innovative research paper formats (e.g. Data Paper, OMICS Data Paper, Software Description, R Package, Species Conservation Profiles, Alien Species Profile), expert-provided data audit for each data paper submission, automated data export and more.
Don’t forget to also sign up for the BDJ newsletter via the Email alert form on the journal’s homepage and follow it on Twitter and Facebook.
***
Earlier this year, Knowledge Pixels and Pensoft presented several routes for readers and researchers to contribute to research outputs – either produced by themselves or by others – through nanopublications generated through and visualised in Pensoft’s cross-disciplinary Research Ideas and Outcomes (RIO) journal, which uses the same nanopublication workflows.

